SLiCE from Escherichia coli laboratory strains
![]() SLiCE (Seamless Ligation Cloning Extract) from Escherichia coli laboratory strains
SLiCE (Seamless Ligation Cloning Extract) from Escherichia coli laboratory strains
| Manipulation of recombinant DNA molecules is an indispensable step in modern
molecular biology. Type IIP restriction endonucleases and DNA ligases were
the original “workhorses” utilized to generate plasmids or other types
of DNA vectors. Recently, various restriction endonuclease cleavage site-independent
cloning methods that overcome the limitations associated with the lack
of unique restriction enzyme sites have been described. These methods are
known as the so-called "seamless cloning", and overlapping sequences
present at the 5′- and 3′-ends of DNA fragments are combined into vector
in vitro. Seamless DNA assembly kits such as Gibson assembly kit and In-Fusion
kit, have become commercially available. However, many commercially available
kits are associated with a high cost per reaction. The SLiCE method can
use extracts from the commonly available Escherichia coli (E. coli) laboratory strains for homemade seamless DNA cloning kit; these extracts
can be easily prepared in the laboratory. This method greatly reduces the
costs associated with DNA manipulation, and can be used as an alternative seamless DNA cloning method to commercially
available kits. |
|||
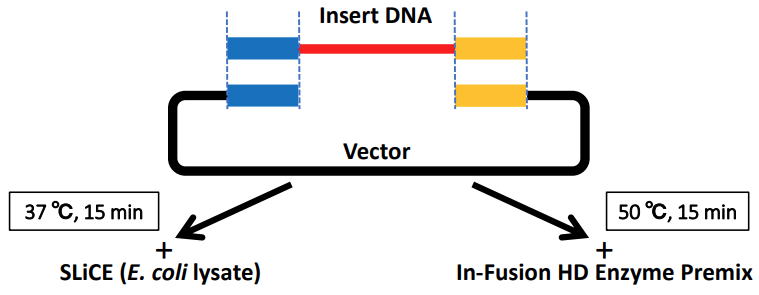 |
|||
| 1. SLiCE from Escherichia coli laboratory strains SLiCE has been initially developed for a novel bacterial cell extract-based DNA cloning method that utilizes the in vitro seamless cloning activity of E. coli cell extracts. Cell extracts of E. coli PPY strain that carries the lambda prophage Red/ET recombination system can efficiently assemble DNA fragments with short-end overlaps in vitro (ref.1). Cell lysates from E. coli RecA- strains such as DH10B also contain endogenous in vitro seamless cloning activity. However, cloning with lysates from these strains had not been efficient, particularly in the case of inserts with short homology lengths (approximately 15-20 bp), because of a lower colony formation rate (ref.1). To extend the utility of this system, I comprehensively surveyed the optimal conditions required for simple and efficient SLiCE cloning by using cell extracts from several commonly used E. coli laboratory strains (ref.2). By incorporating some modifications into the SLiCE preparation protocol and optimizing the reaction conditions, I have obtained SLiCE preparations from several E. coli laboratory strains that yielded good results for seamless assembly of DNA with short end overlaps (approximately 15-19 bp). These SLiCEs can be prepared using commercially available CelLytic B Cell Lysis Reagent (Sigma, B7435) (ref.2) or buffers containing Triton X-100 (ref.3, ref.4). |
|||
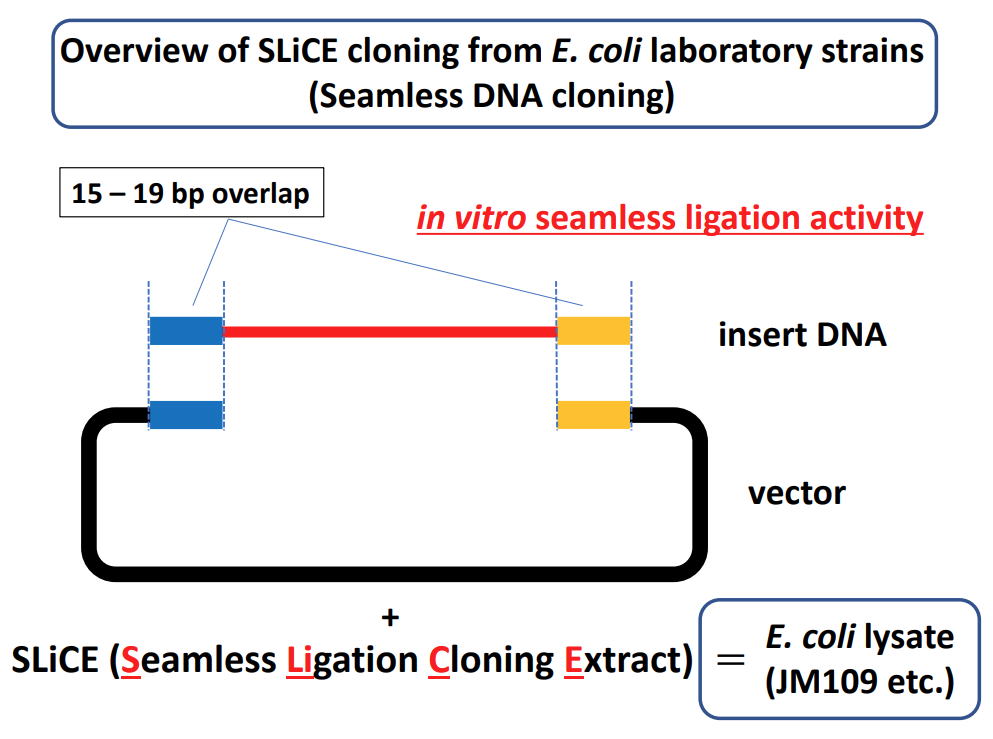 |
|||
| 2. SLiCE-mediated PCR-based (SLiP) site-directed mutagenesis QuikChange site-directed mutagenesis is widely used as a simple polymerase chain reaction (PCR)-based method that does not require purification of PCR products. However, mutations may occur in the vector at non-desired sites during PCR amplification. Such mutation can compromise the fidelity of the approach. To overcome this problem, I demonstrated that SLiCE-mediated PCR-based site-directed mutagenesis (SLiP site-directed mutagenesis) could be performed using the extracts of laboratory E. coli strains (ref. 2). This approach can be adopted in studies that require precise generation of mutants in the absence of unwanted alterations to the vector backbone. ---------------------------------------- |
|||
| 1.Preparation of SLiCE (from JM109) (ref.5) JM109 stored at -80℃ ※DH5a, XL10-Gold and Mach1-T1R strains can also be used. | 1mL LB is maintained at 37℃, usually for 3 h. | 50mL 2xYT is maintained at 37℃, usually for 4-5 h. Harvest the cells at OD = 2 to 3. ※If OD does not reach to 2-3, incubate overnight at 37℃. Next day, transfer 1mL overnight culture into another 50mL 2xYT and incubate at 37℃ for several hours. ※We measure the OD by UV-visible spectrophotometer V-650 (JASCO corp.) Centrifuge at 5,000xg for 10min at 4℃. Wash in 50mL ice-cold Milli-Q water. Centrifuge at 5,000xg for 5min at 4℃. 0.3-0.4 g of cells (wet weight) Resuspend gently in 1.2mL CelLytic B Cell Lysis Reagent (Sigma, B7435) (ref.2). ※Alternatively, buffers containing Triton X-100 can also be used (ref. 3, ref.4). Incubate it for 10min at room temperature. ※Leave to stand, not rotate. Centrifuge at 20,000xg for 2min at 4℃. ※If you want to evaluate efficiency of protein extraction, you should check protein concentration of supernatant (ref. 3). Place the supernatant on ice and add 1 volume of ice-cold 80% glycerol (v/v). Aliquot 40uL of each SLiCE extract into a 0.2mL-PCR tube. Snap-freeze in a bath of liquid nitrogen. Maintain this stock solution at -80℃ (for use, 40uL each extract in 0.2mL 8-strip PCR tube). ※SLiCEs can be stored at -80℃ for at least three years without significant loss of activity. ※96-well paper box for PCR-tube (0.2mLx8) is useful, when SLiCEs can be stored at -80℃. The 96-well papaer box for 0.2mLx8 PCR tube is here. ※For short-term storage, SLiCEs are stored at -20℃. ※SLiCEs can be stored at -20℃ for 2-3 months without significant loss of activity. 2.10 x SLiCE buffer(0.2um filtered), -20℃ stock 500mM Tris-HCl (pH7.5) 100mM MgCl2 10mM ATP 10mM DTT 3.Vector ・Both PCR amplified vector and restriction enzyme-digested vector can be used. ・The presence of a heterologous flanking region at the 5'- or 3'- end of the vector DNA does not inhibit accurate ligation of the insert DNA into the vector, although the presence of heterologous flanking regions at both the 5'- or 3'- ends of the vector DNA reduces cloning efficiency. ・We usually stock several vectors digested with typical restriction enzymes for SLiCE (Some flanking region are available for SLiCE). 4.Insert DNA ・Primers are designed with a 19-bp overlap region (any primer >15bp long should be acceptable). ・Insert DNA fragments should be amplified by a high-fidelity DNA polymerase. 5.Standard protocol ・Perform PCR (19-bp overlap sequence) ・Purify the PCR fragments by using a silica column) If multiple bands are amplified by PCR, the fragments should be purified by agarose gel electrophoresis. ・Vector(10-100ng) + insert DNA (insert : vector=1:1 to 3:1) ※Excess insert DNA inhibits the SLiCE reaction (ref. 4). ・SLiCE reaction Insert DNA Vector DNA 10xB 1uL SLiCE 1uL add DDW up to 10uL 37℃ for 10-60 min (prolonged incubation reduces colony formation rate). ※A SLiCE reaction for 15 min results in enough cloning-efficiency, in many cases. 6.Rapid protocol ・Perform PCR (19-bp overlap sequence) ・Vector(10-100ng) + unpurified insert DNA 1uL ・SLiCE reaction Unpurified insert DNA 1uL ※DpnI treatment efficiently reduces background colony formation. ※When a restriction enzyme-digested vector is used, DpnI should be inactivated for 15 min at 80℃. Vector DNA 10xB 1uL SLiCE 1uL add DDW up to 10uL 37℃ for 15min 7.SLiCE-mediated PCR-based (SLiP) site-directed mutagenesis (ref.2, ref.5) ・Design of mutation primers by PrimerX Condition1: melting temperature >78℃ Condition2: primer termination in G or C for QuikChange site-directed mutagenesis kit. ・Two fragments containing a site-directed mutation site should be amplified. ・Rapid SLiCE protocol Unpurified fragment A 1uL Unpurified fragment B 1uL ※DpnI treatment efficiently reduces background colony formation. ※When a restriction enzyme-digested vector is used, DpnI should be inactivated for 15 min at 80℃. Vector DNA 10xB 1uL SLiCE 1uL add DDW up to 10uL 37℃ for 15min If necessary, PCR fragments can be purified and reacted using the standard SLiCE protocol. ---------------------------------------- SLiCE-related FAQ (Frequently Asked Questions) We have received many questions about SLiCE from laboratory E. coli strains, so far. Among them, some representative questions are summarized as FAQ. Q1. Which E. coli strain do you recommend for SLiCE-preparation? A1. Various laboratory E. coli strains (JM109, DH5a, XL10-Gold, Mach1-T1R etc.) can be used as a source of SLiCE. We usually prepare SLiCE from JM109 because that showed a slightly higher colony formation rate (ref.2, Table 1). Q2. How much volume of bacterial culture do you recommend to prepare SLiCE? A2. In our experience, the quantity of SLiCE prepared from 50 mL E. coli culture was sufficient to perform over two thousand SLiCE reactions, which implies that once prepared, it can be used for several years. Q3. How long does it take to prepare SLiCE? A3. SLiCE can be prepared in a day. Only four solutions, namely bacterial medium, lysis buffer, MilliQ, and 80 % glycerol, need to be prepared and kept ready the day before. Either a commercially available lysis buffer or a Tris-buffer containing TritonX-100 may be used as the lysis buffer here. Preparation of SLiCE is easier than preparation of chemically competent E. coli cells. Q4. When E. coli was cultured according to your protocol for SLiCE-preparation, the culture did not reach OD = 2–3 even at night. What would be the best thing to improve the growth conditions? A4. Please cultivate overnight, in accordance with the protocol. We recommend that 1 mL of overnight-grown culture be transferred to 50 mL of fresh medium, next morning. Q5. In case the E. coli culture went over OD = 3, what would be the best thing to do? A5. We recommend that 1 mL of the over-grown culture (50 mL) be transferred to 50 mL of fresh medium. Q6. How do you store E. coli SLiCEs for long time? A6. SLiCEs can be stored at -80 ℃ for at least five years, without significant loss of activity. For short-term storage, say for 2–3 months, SLiCEs may be stored at -20 ℃ without significant loss of activity. We usually replace the SLiCE, at -20 ℃, with a new SLiCE stock after two months. When SLiCEs are stored at -20 ℃, we recommend using a cooler rack for the storage of restriction enzymes to maintain the temperature. We have a success case in cloning of a single fragment, using SLiCE that was stored at -20℃ for 5.5 months, although the cloning of a single fragment showed low efficiency . Q7. Which is a more efficient cloning method, restriction enzyme-method or SLiCE-method? A7. SLiCE, which is a seamless DNA cloning method, shows higher cloning efficiency (ref.2, ref.4), and can simultaneously incorporate multiple DNA fragments into a vector (Figure 1 and Figure 2). Q8. Can we use SLiCE as an alternative to commercially available seamless DNA cloning kits? A8. Definitely, SLiCE from laboratory E.coli strains can serve as an alternative to commercially available kits (ref.4). Q9. What length of homologous DNA region do you recommend? A9. We recommend a 19-bp overlap in the short-end homology regions between insert and vector DNAs because, in our experience, 19-bp overlap sequences showed a relatively higher colony formation rate (ref.2, Table 2). However, 15-bp overlap sequences also work with considerably good efficiency (ref.3, ref.4). Q10. How do you prepare the vector backbone? A10. Both PCR-amplified vector and restriction enzyme-digested vector are suitable with comparable efficiency. Multiple flanking regions in the restriction enzyme-digested vector may reduce the colony formation rate, although they do not inhibit accurate ligation of the insert DNA into the vector (ref.2, Table 3). We usually stock several vectors, digested with typical restriction enzymes, for SLiCE. Q11. Do you recommend purification of PCR-amplified fragments for SLiCE-reaction? A11. Unpurified DNA inserts can be ligated to vector DNA, but with reduced efficiency (ref.2, Table 4). Therefore, we recommend the purification of insert DNAs by a Gel/PCR purification kit (i.e. FastGene Gel/PCR Extraction Kit). Q12. PCR products, treated with Cloning Enhancer in In-Fusion system, can directly be applied to In-Fusion reaction. It is very useful. Can a similar process be applied to SLiCE-method? A12. ExoSAP-IT (Thermo Fisher Scientific) can be applied to SLiCE method, similar to the process mentioned. When PCR products are treated with ExoSAP-IT at 37 ℃ for 15 min and at 80 ℃ for another 15 min, they can be directly applied to the SLiCE reaction. If the template DNA, contained in the PCR products, need to be removed, DpnI may be added together with ExoSAP-IT. However, the enhancement of cloning efficiency by ExoSAP-IT treatment (30 min) is limited (ref.2, Table 4). Alternatively, purification by a Gel/PCR purification kit (15 min) shows a higher cloning efficiency (ref.2, Table 4), consuming less time than the ExoSAP-IT treatment, although involving multiple operations. Q13. What is the cloning efficiency (the ratio of correct clones)? A13. In SLiCE cloning, colony PCR is not required for screening positive clones, because almost all the colonies contain plasmids with the PCR-amplified fragment incorporated (ref.2; Table 1 and 2, and ref.3; Table S1). Q14. When E. coli cells are transformed with SLiCE reaction solution, does recovery culture using SOC medium improve transformation efficiency? A14. Recovery culture of E. coli cells improves transformation efficiency of SLiCE-method, regardless of the type of antibiotic. We strongly encourage you have a time for recovery culture of E. coli cells. Q15. How much competence of E. coli cells is required for SLiCE cloning? A15. We generally chemically competent E. coli cells ~107 CFU for SLiCE cloning. Generally, E. coli competent cells (~106 CFU) have enough competence for cloning of one DNA fragment. Q16. Can SLiCE clone multiple DNA fragments into a vector, simultaneously? A16. Yes, two DNA fragments can be assembled and inserted simultaneously into a vector, because SLiCE is a highly efficient seamless cloning method (Figure 1). Moreover, each of the two DNA fragments can be simultaneously inserted into two different sites in a vector (Figure 2). We also clone three DNA fragments into a vector, simultaneously. In this case, we recommend to use a new SLiCE from a stock at -80℃. Q17. We failed cloning of small DNA fragments in SLiCE-method. Is SLiCE-method an application for larger DNA fragments cloning? A17. SLiCE-method can be applied to the cloning of DNA fragments of various sizes. In our experiences, we could insert 42 bp DNA sequence into the vector by SLiCE-method. However, in several cases, we detected multiple tandem insertion of DNA fragments into a vector site when small DNA fragments were applied. In this case, quantity of insert DNA fragments should be reduced in SLiCE-reaction. We recommend molar ratio of i:v = 1:1 to 3:1 in SLiCE-reaction. ※According to the report by other group, they could insert the 90 bp DNA sequence into the vector. Q18. Are the insert and vector DNA fragments degraded by nuclease which may be contained in the E. coli lysate? A18. SLiCE may include some DNA degradation activities from E. coli cells. To prevent any undesirable DNA degradation, we recommend the following: (a) SLiCE reaction should not be incubated for a long time (usually 15 min at 37 ℃), (b) after reaction, the SLiCE solution should be either immediately transformed to chemically competent cells or immediately frozen. Q19. How much does SLiCE-preparation cost? A19. SLiCE method is a cheaper approach than either the method employing DNA ligase or the commercially available seamless DNA cloning kit (ref.4, ref.5); cost per reaction: 0.4 Yen (> 50 % of this cost is attributed to the price of plastic tubes). Q20. Recently, it was reported that in in vivo E. coli cloning (iVEC)-method, vector and multiple DNA fragments can be assembled using in vivo homologous recombination activity in E. coli cells. Why would you recommend using SLiCE that requires additional in vitro reaction? A20. It is commonly known that if both linear insert and vector DNA are directly introduced to E. coli cells, they can be assembled using in vivo homologous recombination activity of E. coli cells. In contrast, the SLiCE method utilizes E. coli homologous recombination activity, in vitro. At short-end homology region (15-19 bp), the cloning efficiency of SLiCE method was found to be one or two orders of magnitude higher than that of the method using endogenous in vivo homologous recombination activity (ref.2 Table 1, "JM109" vs "no extract", ref.6). Q21. What are the differences between QuikChange-method and SLiP-method? What is the merit in SLiP-method? A21. QuikChange-method amplifies the full-length plasmid, containing both insert and vector regions, by PCR. In contrast, SLiP-method amplifies only the insert regions by using a restriction enzyme-digested vector. Hence, PCR-time gets reduced in SLiP-method, because it amplifies only the insert DNA. The procedure of SLiP site-directed mutagenesis is comparable to that of QuikChange site-directed mutagenesis, in terms of simplicity and efficiency, because SLiCE can use unpurified PCR fragments as well. Furthermore, the possibility of introducing a mutation into the vector region is excluded by the use of a restriction enzyme-digested vector in SLiCE cloning. Q22. How long does a conventional cloning by SLiCE method take? A22. We usually carry out SLiCE-cloning in the following schedule. Day 1 14:00 Primers delivered. 14:30 PCR performed 16:30 Check for PCR-amplification, followed by DNA fragment-purification 1/10 volume: PCR-amplification check by agarose electrophoresis 9/10 volume: DNA fragment-purification by FastGene Gel/PCR Extraction Kit (Nippon Genetics) ※The two above-mentioned tasks are executed in parallel. 17:00 SLiCE (37℃, 15 min) ※Restriction enzyme-digested vectors are stocked in laboratory, in advance. 17:30 Transformation 19:00 Plating on LB plate. ※The subsequent steps are performed even if a PCR-amplified band is not detected on agarose gel. ※We often get correct clones, even if we cannot detect PCR-amplification on gel by 1/10 volume. Day 2 9:00 Screening (Colony-PCR or plasmid preparation from 2 mL LB medium) |
|||
| ref.1) | Zhang, Y. et al., | Nucleic Acids Res. vol.40, e55 (2012). | |
| ref.2) | Motohashi, K., | BMC Biotechnol. vol.15, 47 (2015). | |
| ref.3) | Okegawa, Y. and Motohashi, K., | Anal. Biochem. vol.486, 51-53 (2015). | |
| ref.4) | Okegawa, Y. and Motohashi, K., | Biochem. Biophys. Rep. vol.4, 148-151 (2015). | |
| ref.5) | Motohashi, K., | Methods Mol. Biol. vol.1498, 349-357 (2017). | |
| ref.6) | Motohashi, K., | Biochem. Biophys. Rep. vol.9, 310-315 (2017). | |
| ---------------------------------------- More information is provided in the following references. Please cite these references (ref. 2, 4, 5), if you use the SLiCE from laboratory Escherichia coli strains for your scientific work. |
|||
| ref.2 | Ken Motohashi | ||
| "A simple and efficient seamless DNA cloning method using SLiCE from
Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis" BMC Biotechnol. 15, 47 (2015) |
|||
| ref.3 | Yuki Okegawa and Ken Motohashi | ||
| "Evaluation of seamless ligation cloning extract (SLiCE) preparation
methods from an Escherichia coli laboratory strain" Anal. Biochem. 486, 51-53 (2015) |
|||
| ref.4 | Yuki Okegawa and Ken Motohashi | ||
| "A simple and ultra-low cost homemade seamless ligation cloning extract
(SLiCE) as an alternative to a commercially available seamless DNA cloning
kit" Biochem. Biophys. Rep. 4, 148-151 (2015) |
|||
| ref.5 | Ken Motohashi | ||
| "Seamless Ligation Cloning Extract (SLiCE) method using cell lysates
from laboratory Escherichia coli strains and its application to SLiP site-directed mutagenesis" Methods Mol. Biol. 1498, 349-357 (2017) |
|||
| ref.6 | Ken Motohashi | ||
| "Evaluation of the efficiency and utility of recombinant enzyme-free
seamless DNA cloning methods" Biochem. Biophys. Rep. 9, 310-315 (2017) |
|||
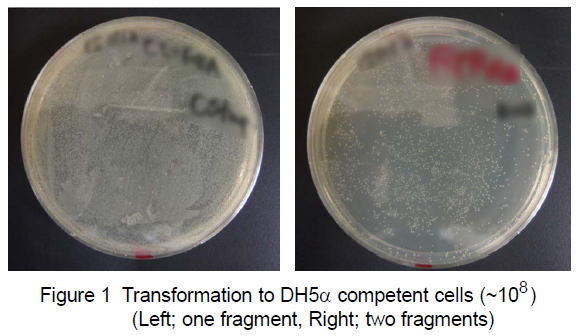 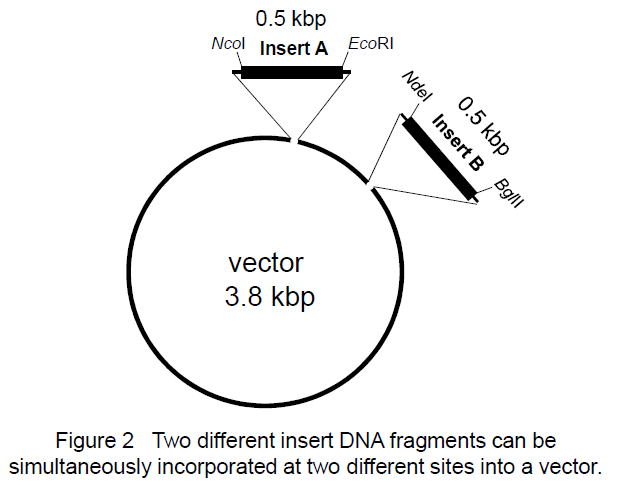 |
|||
 |
||||
| Return to "Technical tips" | ||||
| E-mail: |