Pattern formation in two dimensions
Stanislav Y. Shvartsman
Departments of Chemical and Biological Engineering and Molecular Biology; Lewis-Sigler Institute for Integrative Genomics, Princeton University.
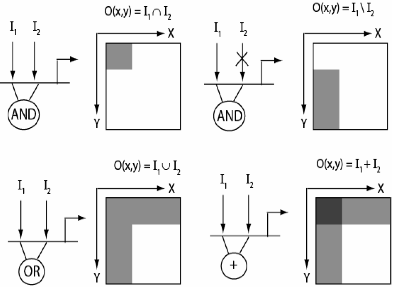
During Drosophila oogenesis, a gradient of Epidermal Growth Factor Receptor (EGFR) activation patterns the follicle cells, which form an epithelium over the developing oocyte. In response to their patterning by EGFR, follicle cells acquire a number of different fates necessary for the formation of the eggshell, a complex structure that protects the embryo and mediates its interaction with the environment. Genetic studies of Drosophila oogenesis established it as a premiere model for the mechanistic analysis of developmental signaling [1]. Based on the results of these studies, we are using the EGFR-patterning of the follicle cells to establish systems-level models of pattern formation by locally activated signaling pathways. We have quantified the gradient of EGFR activation in the follicle cells, identified dozens of its target genes, and begun to explore mechanisms of their regulation [2-7]. Analysis of the expression patterns of these genes suggests that they obey a combinatorial code, based on a small number of building blocks and geometric operations (Figure 1). The building blocks in this code are related to the distributions of inductive signals that pattern the follicular epithelium. The geometric operations can be potentially explained by the Boolean operations at the cis-regulatory modules of the EGFR target genes. Our recent experiments identified some of these modules, providing support for the combinatorial code hypothesis.
1.Berg, C.A. (2005). The Drosophila shell game: patterning genes and morphological change. Trends Genet 21, 346-355.
2.Goentoro, L.A., Reeves, G.T., Kowal, C.P., Martinelli, L., Schupbach, T., and Shvartsman, S.Y. (2006). Quantifying the Gurken morphogen gradient in Drosophila oogenesis. Dev Cell 11, 263-272.
3.Yakoby, N., Bristow, C.A., Gong, D., Schafer, X., Lembong, J., Zartman, J.J., Halfon, M.S., Schüpbach, T., and Shvartsman, S.Y. (2008). A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell 15, 725-737.
4.Lembong, J., Yakoby, N., and Shvartsman, S.Y. (2009). Pattern formation by dynamically interacting network motifs. Proc Natl Acad Sci 106, 3213-3218.
5.Yakoby, N., Lembong, J., Schupbach, T., and Shvartsman, S.Y. (2008). Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development 135, 343-351.
6.Zartman, J.J., Kanodia, J.S., Cheung, L.S., and Shvartsman, S.Y. (2009). Feedback control of the EGFR signaling gradient: superposition of domain splitting events in Drosophila oogenesis. Development 136, 2903-2911.
7.Zartman, J.J., Cheung, L.S., Niepielko, M., Bonini, C., Haley, B., Yakoby, N., and Shvartsman, S.Y. (2011). Pattern formation by a moving morphogen source. Physical Biology 8, 045003.